Abstract
Introduction
Acute myeloid leukemia (AML) represents 20% of pediatric leukemias. Despite therapeutic improvements, ∼30% of children die due to therapy-related toxicity and relapse. In solid tumors, the density and functional orientation of the tumor microenvironment (TME) influence prognosis and response to chemotherapy and immunotherapy. Defining immunologic features associated with unfavorable response to standard treatment in pediatric AML might improve stratification systems that can be used for experimental interventions and personalized approaches. In this study, we investigated the potential mechanisms and effect of immune response and evasion in bone marrow (BM) of AML pediatric patients through an integrative gene expression analysis.
Methods
Thirty-two BM aspirates were collected from pediatric patients with non-promyelocytic AML at diagnosis in 3 different hospitals in Qatar, Pakistan, and Italy. Patients were assigned to 2 prognosis-related groups: patients who maintained a complete remission (CR) after first line of chemotherapy (sustained CR) vs patients who progressed/relapsed and/or obtained CR after hematopoietic stem cells transplantation (HSCT) (non-sustained CR). BM mRNA samples were analyzed with RNA sequencing (RNA-Seq) and/or NanoString's PanCancer IO 360 Gene Expression research panel. In particular, mRNA of 25 samples (Cohort 1, discovery set) were analyzed by NanoString's PanCancer IO 360. Subsequently, a subset of samples from Cohort 1 (n=19), was analyzed by RNA-Seq for technical validation. Cohort 1 results were then used to build a predictive classifier that was tested on Cohort 2 samples (n=7, validation set), analyzed with RNA-Seq.
Results
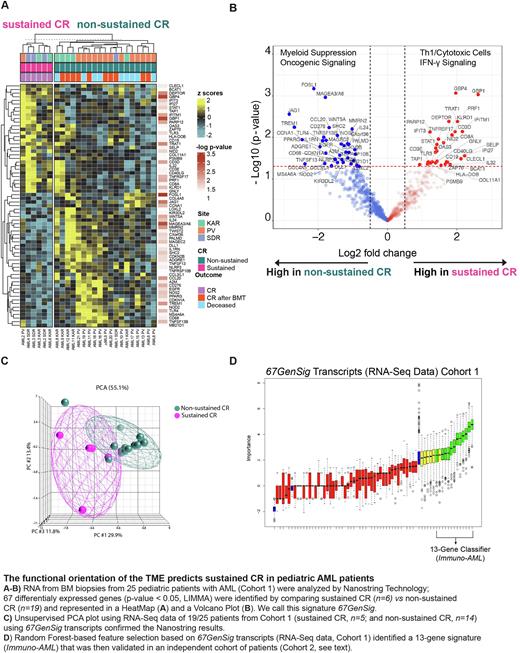
Gene expression analysis of 25 samples (Cohort 1, sustained CR, n=6; non-sustained CR, n=19) using NanoStringassay revealed 67 significant differentially expressed genes (DEGs; p < 0.05, LIMMA) among sustained CR vs non-sustained CR groups. Patients with sustained CR displayed coherent overexpression of immune genes related to IFN activation (IFIT3, IFITM1, IFI27, STAT1, GBP1), cytotoxic function (PRF1, GNLY), antigen presentation and B cell functions (TAP1, CD19), and T cell infiltration (CD3D, CD8A), consistent with a favorable T helper 1 (Th1)-cytotoxic polarization of the TME. This pattern was not observed in non-sustained CR patients, which overexpressed genes consistent with macrophage-myeloid suppression (CD68, TREM1) and intrinsic oncogene signaling linked with T cell exclusion and immune suppression (WNT5A, WNT-β- catenin pathway, and EFGR) (Fig1 A,B). This 67-gene signature (67GenSig) was technically validated using an orthogonal platform (RNA-Seq) on a subset of 19 patients (sustained CR, n=5; non-sustained CR, n=14) for which sufficient RNA-Seq data was available (65 out of 67 transcripts were available for analysis, 2 transcripts were filtered out after QC). Unsupervised PCA (Fig1 C) based on 67GenSig clearly segregated samples according to outcome, confirming the previous NanoString results.
Next, we sought to identify a predictive classifier to precisely discriminate sustained CR from non-sustained CR AML pediatric patients using a minimal number of genes. By applying a Random Forest-based feature selection on 67GenSig transcripts of RNA-Seq data from Cohort 1 samples, we developed a 13-gene classifier (Immuno-AML) that was tested in a new independent dataset (Fig1 D; Cohort 2, n=7: sustained CR, n=4; non-sustained CR, n=3). Overall, genes included in the Immuno-AML predictor substantiated a positive Th1/cytotoxic vs myeloid polarization. The Immuno-AML predictor was able to classify patients in the validation cohort (Cohort 2) in "sustained CR" and "non-sustained CR" prognostic groups with a very good performance achieving 85.71% accuracy, 100% specificity, and 75% sensitivity.
Conclusion Our data suggest that response to conventional therapy in AML pediatric patients is conditional to the presence of a Th1 polarization of the TME while is prevented by myeloid-mediated immune suppression and intrinsic oncogenic signaling. The optimized predictive signature (Immuno-AML) might be tested and/or refined in larger cohort to develop a clinical-grade predictive biomarker. It might help to provide a rational guidance for risk stratification criteria and personalized treatment decision paths, implying the possibility of combining immunotherapy drugs.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal